Our Science
Critical insights and better biomarkers through RNA analysis
DNA mutational analysis and other traditional approaches for identifying responders fall short in predicting all patients who will respond to a specific therapy. Other functional drivers of response appear buried deep within tumor biology “dark matter.” Although DNA has produced profound precision medicine solutions, and will continue to be important, the low-hanging fruit has been picked and there is a need for data that more dynamically possesses the biological range to reflect the complexities of response.
In contrast to static genetic mutations, RNA reflects the phenotype, which is the inherent expressed traits of a tumor encompassing cancer cells, their associated genetic drivers, collaborating entities such as stroma, and the immune environment. This collective ecosystem is the tumor and the immune microenvironment (T(I)ME).
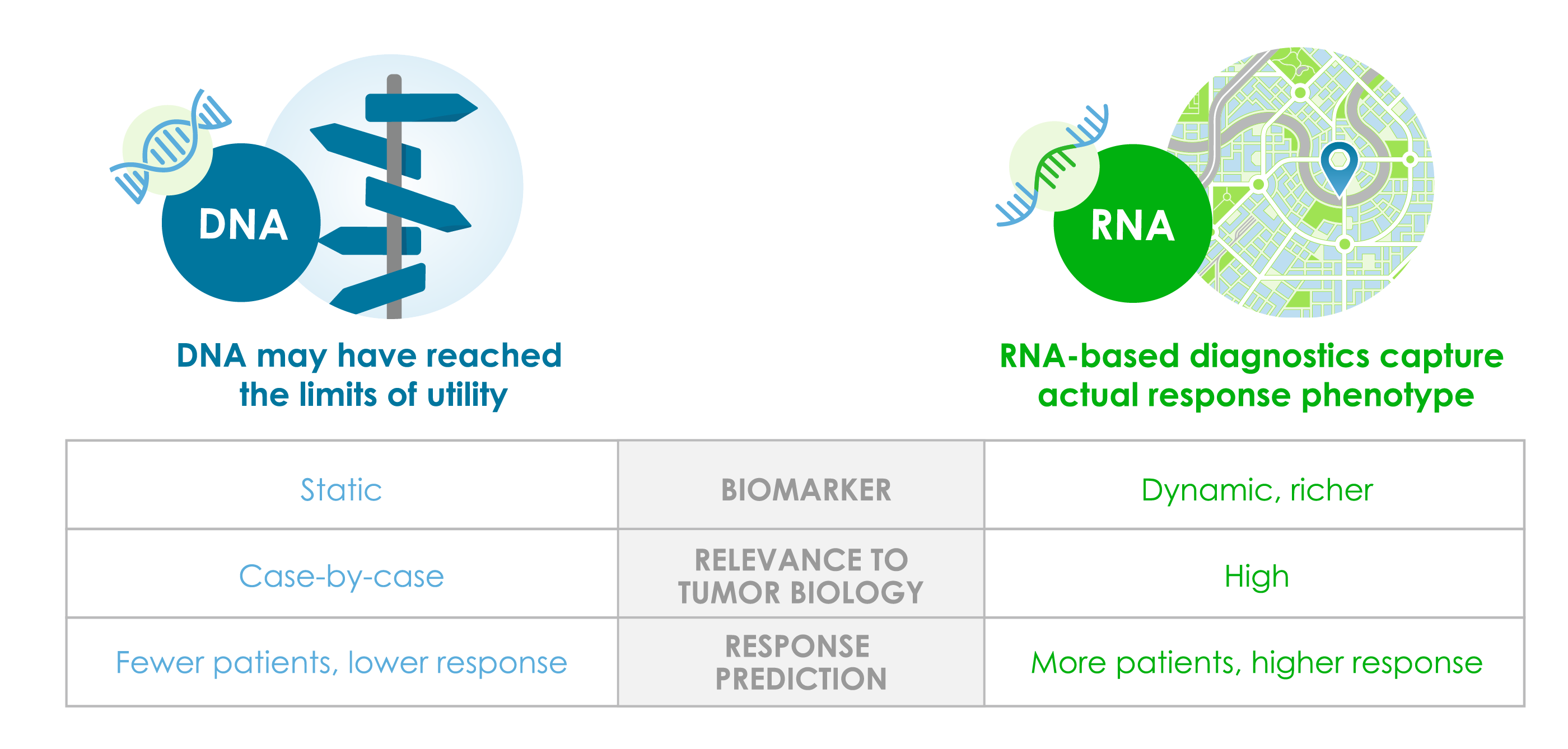
Deciphering this complex ecosystem to shed light on the biological “dark matter” of the tumor is expanding precision medicine solutions. Our platform offers an innovative, more systematic RNA-based approach that may better pinpoint treatments and ultimately have the most positive impact on patient outcomes.
Clinical need for drug response biomarkers
Today, new treatment approaches such as immunotherapy and other molecular-targeted therapies are changing the landscape for cancer treatment. This has resulted in dramatic improvements in long-term survival rates compared with conventional chemotherapy, but the benefit has only been realized in a subset of patients.
At the same time, many promising cancer therapeutics are failing in late-stage clinical trials. However, pronounced effects seen in small numbers of study participants suggest that by better identifying responders we could drive greater success.
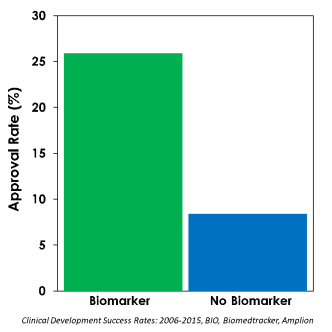
Recent pivotal clinical trials confirm this with higher approval rates (25.9%) for drugs developed using response-identifying biomarkers compared to drugs without such biomarkers (8.4%). (Clinical Development Success Rates: 2006-2015, BIO, Biomedtracker, Amplion)
Clinical evidence
At this time, established biomarkers tied to specific drug mechanisms or disease pathways have not been widely successful or consistently reliable in identifying responder populations. Many cancers have proven too complex and heterogeneous. For example, PD-L1 levels have not proven to be a consistent predictor of PD-1 inhibitor efficacy in the immunotherapy context, suggesting that the responder profile is more complex than expression of a single molecule. (Nivolumab in First Line or Recurrent Non-Small Cell Lung Cancer, Carbone, D.P., NEJM, June 22, 2017.)
Similarly, many targeted therapies have produced significant responses in patients where the drug’s target is not altered or (over) expressed or failed to benefit patients with an alteration predicted to deliver a response. For example, EGFR-directed therapies have demonstrated efficacy in patients lacking EGFR mutations. (Zhu CQ, da Cunha SG, Ding K, et al: Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol 26:4268-4275, 2008.)
Even in indications with dense coverage of the tumor mutational landscape, such as non-small cell lung cancer (NSCLC), a large proportion of patients have no identifiable precision medicine options due to lack of accompanying biomarkers. (Pakkala, Suchita, and Suresh S. Ramalingam. “Personalized therapy for lung cancer: striking a moving target.” JCI insight 3.15, 2018.)
Our rT(I)ME Explorer Platform boosts drug development
The union of pioneering innovations, proprietary pipelines, AI and machine learning comprises our rT(I)ME Explorer Platform – an integrated set of RNA-based pipelines, signatures and algorithms for building robust RNA gene expression signatures. The platform enhances drug development by unleashing the informative value of RNA as impactful biomarkers, diagnostics or companion diagnostics.
Learn MoreA growing portfolio of RNA diagnostics for cancer
By applying the rT(I)ME Explorer Platform to specific types of cancer and drug response settings, GeneCentric has developed and is expanding its pipeline of cancer subtypes and biomarkers for some of most prevalent and complex cancers and areas of target biology. Our portfolio across numerous tumor types and target biology is immediately deployable for drug development programs.
Learn More