One sample. Two dimensions. Unlimited potential.
ctDNA isn’t just a vehicle for detecting mutations – it also holds the fingerprint of gene expression. Our GenomicsNext™ platform reads that fingerprint.
How it works:
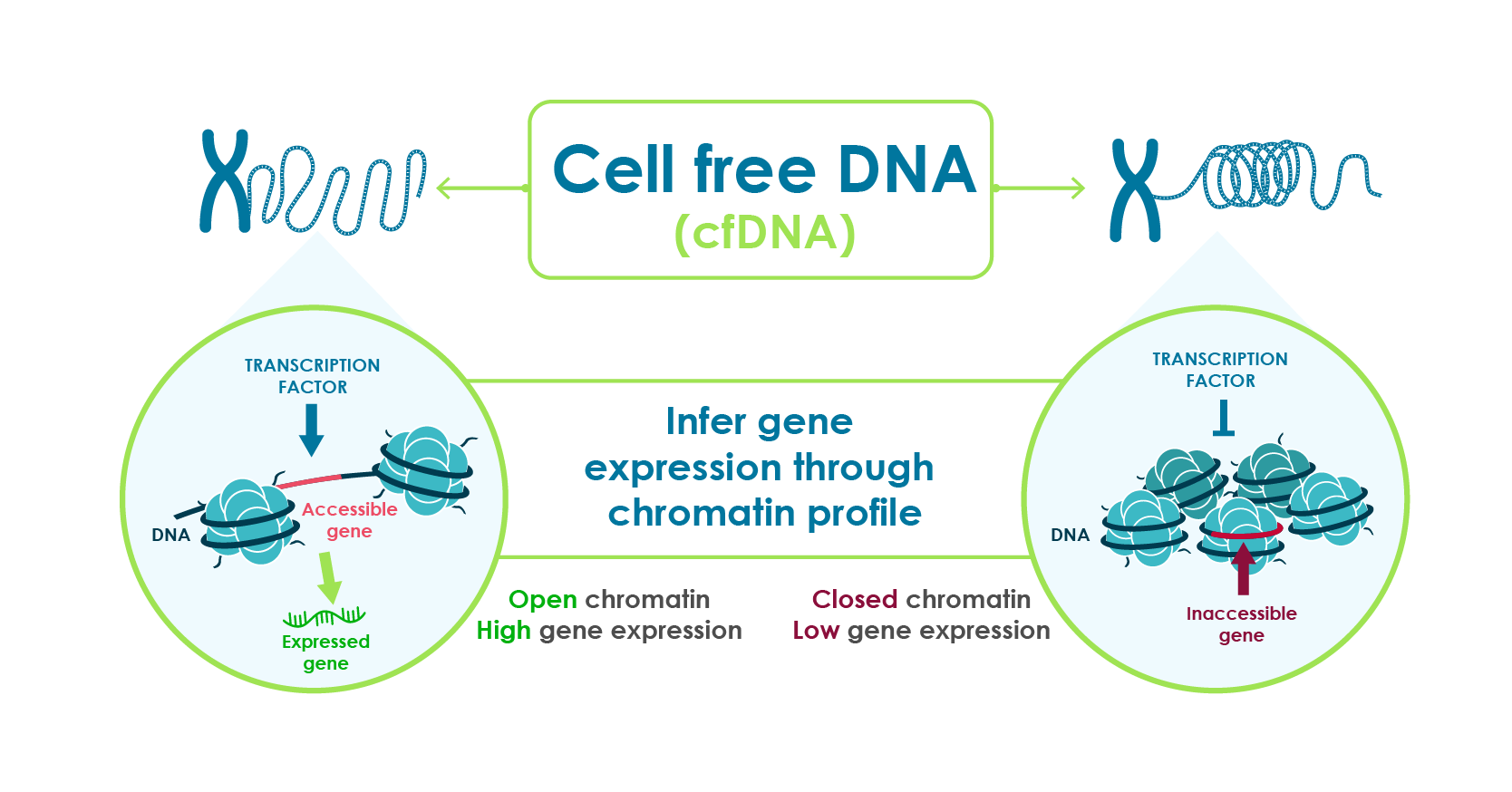
- Actively transcribed regions of DNA have open chromatin that fragment differently during cell death.
- DNA in blood is exposed to natural DNA degrading enzymes and the resulting fragment remnants reveal genomic regions with open chromatin.
- These fragmentation patterns, captured through standard DNA sequencing, reflect the expression activity of genes.
- Our ExpressCT™ algorithms, trained on well-characterized tissue gene expression signatures, sift through noise to extract meaningful expression traits from ctDNA.
Why it matters:
- No RNA required – GenomicsNext uses existing ctDNA workflows
- Integrates seamlessly into standard NGS pipelines
- Scalable and practical for clinical labs and translational research
Capabilities:
- Detect DNA alterations (SNVs, Indels, CNVs, fusions, MSI, TMB)
- Infer gene expression traits (HER2/ER status, subtypes, etc.)
- Build multimodal biomarkers for therapy selection, prognosis, and MRD
- Support ADC development, screen candidates for target expression, and enrich clinical trials
Not all fragments are useful for inferring expression – ExpressCT cancels noise
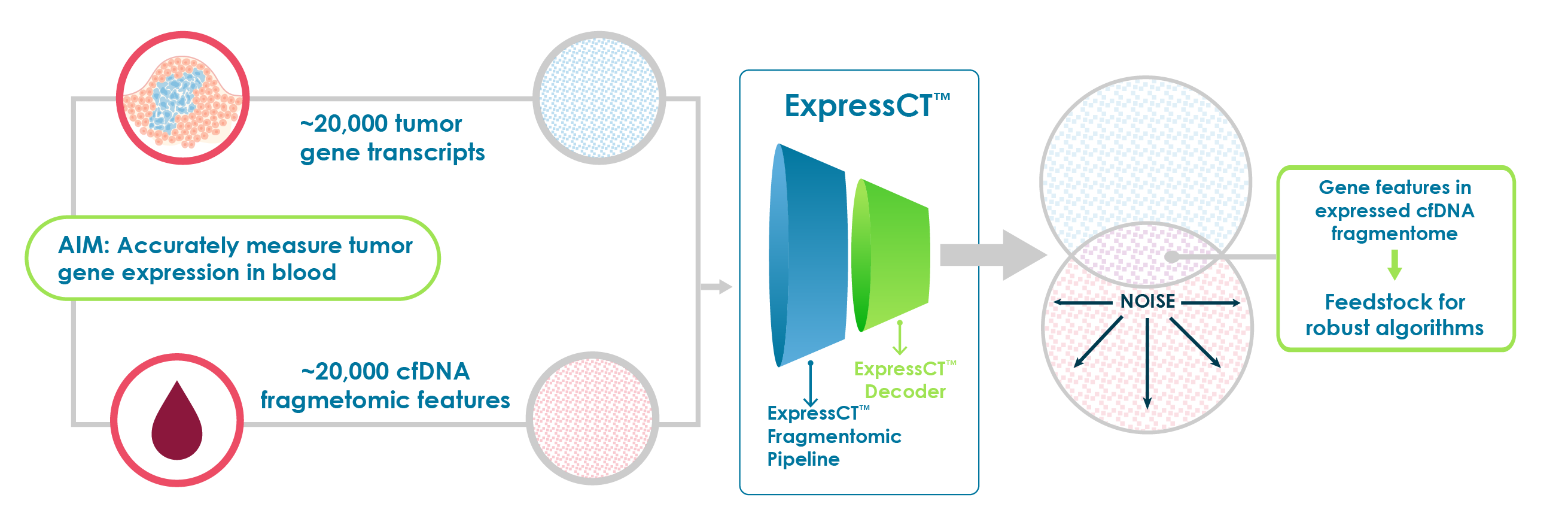
Until now, no sophisticated solution has been available to find ctDNA features that faithfully reflect tumor expression traits. Our ExpressCT technology provides answers to challenges that have prevented integration of gene expression into alteration testing in ctDNA.
We relied on years of experience and a pipeline of RNA-based tissue gene expression signatures to approach the challenge of translating tissue signatures into blood-based versions.
We quickly realized that most ctDNA data was noise. Noisiness and poor sensitivity of gene expression signals make direct one-to-one transfer of gene tumor expression signatures to blood for expression comparison unreliable.
Using advanced, proprietary machine learning, ExpressCT sifts through blood gene expression features to eliminate noise and identify an enriched population of highly concordant ctDNA features that accurately reflect tumor gene expression – the expression-relevant ctDNA fragmentome.
The expression-relevant ctDNA fragmentome can be used to create blood versions of any tumor RNA-based expression signature or create new expression signatures using the expression-relevant ctDNA fragmentome only.
Tumor tissue gene expression signatures
GeneCentric has trailblazed the development of innovative tissue RNA-based gene expression signatures and accumulated deep clinical utility for signatures in pancreatic cancer (PurIST®), bladder cancer (FGFR activation), lung cancer (chemotherapy sensitivity prediction), head and neck cancer (treatment escalation), and colorectal cancer (immuno-oncology sensitivity in microsatellite stable tumors).
| Test Name | Cancer Type | Key Highlights (Reference) |
|---|---|---|
|
PurIST® |
Pancreatic ductal adenocarcinoma |
Identifies molecular subtype of patients with prognostic and predictive survival differences for choosing between first-line standard-of-care options (PMID: 31754050, Wenric S, medRxiv, 2023.) |
|
FGFR activation |
Bladder cancer |
Identifies 2-3x more patients for FGFR inhibitor eligibility compared to DNA tests and prognostic differences have important diagnostic utility (PMID: 39325014) |
|
Adenocarcinoma subtyper |
Non-small cell lung cancer |
2-3x progression-free survival benefit for patients with bronchioid subtype treated with pemetrexed (PMID: 37233991) |
|
Histoplus |
Lung cancer |
Accurately distinguishes lung adenocarcinoma and lung squamous carcinoma and guides treatment selection (PMID: 23701907) |
|
Head and Neck subtyper |
Head and neck cancer |
Identifies molecular subtypes of patients with risk that indicates consideration for treatment escalation (PMID: 36147910) |
|
Microsatellite stable (MSS) predictive response signature |
Colorectal cancer |
Efficiently sorts MSS patients with and without DNA damage and immune gene expression profiles suggestive of responsiveness to combination immuno-oncology therapies (Cancer Res (2024) 84 (6_Supplement): 1040) |
Like PurIST, which is offered by Tempus through a licensing agreement with GeneCentric, tumor tissue gene expression signatures can be readily implemented as a clinical algorithm on any RNA sequencing platform. Liquid biopsy versions of these tests also can be created using ExpressCT technology.
